CROssBAR Use-case: COVID-19 Knowledge Graphs
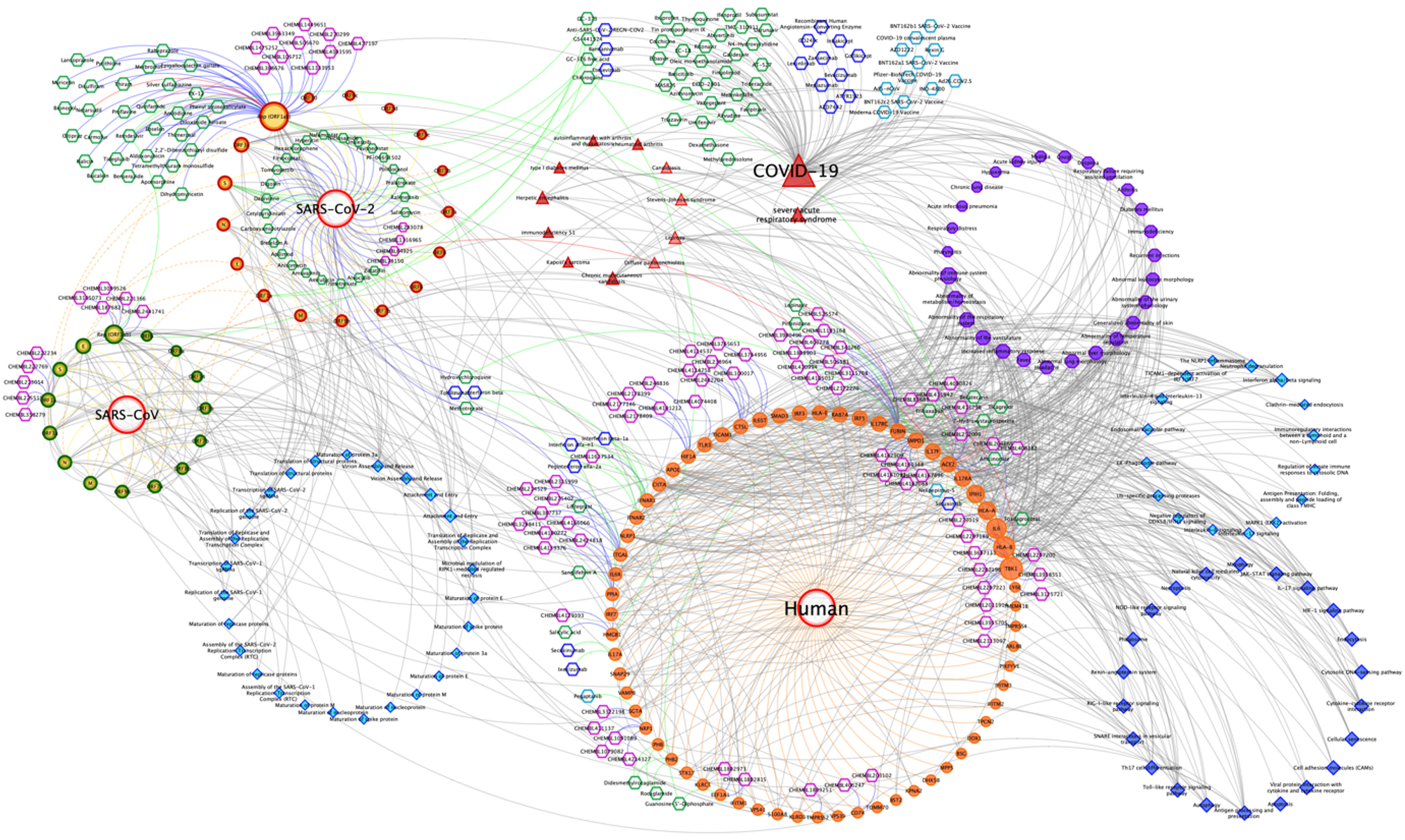
As a use case of the CROssBAR system, we present the COVID-19 / SARS-CoV-2 molecular interactions knowledge graph. We have constructed 2 different versions of the COVID-19 knowledge graph, (i) a large-scale version including nearly the entirety of the related information on different CROssBAR-integrated data sources, which is ideal for further network or machine learning based analysis, and (ii) a simplified version distilled to include only the most relevant human genes/proteins as provided in UniProt-COVID-19 portal ( https://covid-19.uniprot.org) and other related terms, which is ideal for fast interpretation.
The finalized large-scale COVID-19 KG includes 1289 nodes (i.e., genes/proteins, drugs/compounds, pathways, diseases/phenotypes) and 6743 edges (i.e., various types of relations). The simplified COVID-19 KG includes a total of 435 nodes and 1061 edges. Since most of the COVID-19 related data has still not been integrated into the regular pipelines of our source biological databases, the entirety of the data could not be automatically pulled to the CROssBAR database (as of March 2021). As a result, we manually obtained the data from these resources. We applied the same knowledge graph construction methodology incorporated into the CROssBAR system and saved the pre-constructed graphs, which are accessible through the links below.
For more information about the COVID-19 knowledge graphs, please refer to the CROssBAR article or visit the CROssBAR project GitHub repository at: https://github.com/cansyl/CROssBAR