CROssBAR Project
CROssBAR: Comprehensive Resource of Biomedical Relations with Deep Learning Applications and Knowledge Graph Representations
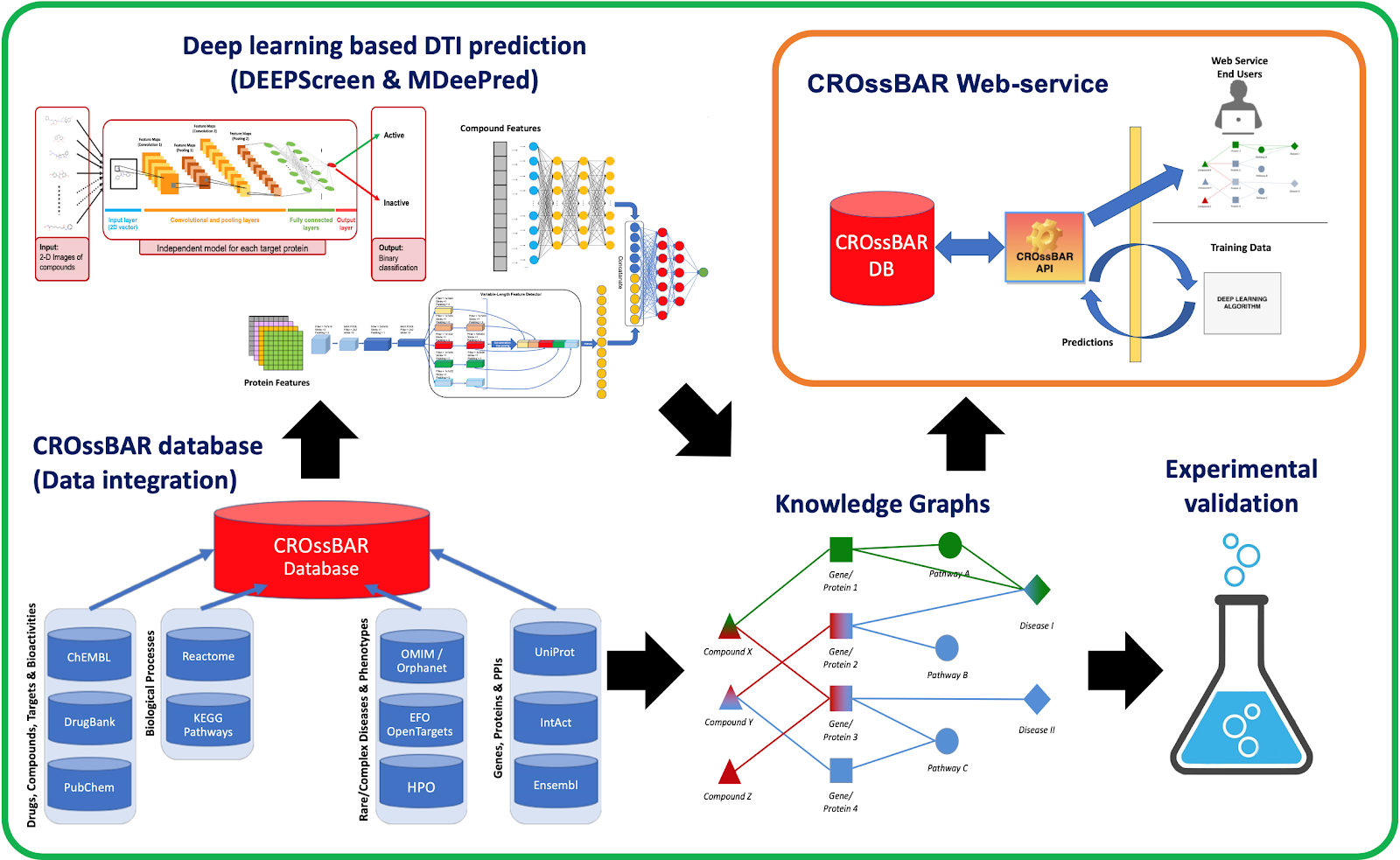
CROssBAR is a comprehensive system that integrates large-scale biomedical data from various resources and store it in a new NoSQL database, enrich this data with deep learning based prediction of relations between numerous biomedical entities, rigorously analyse the enriched data to obtain biologically meaningful modules and display them to the user via easy to interpret, interactive and heterogenous knowledge graphs within an open access, user-friendly and online web-service at https://crossbar.kansil.org.
5 main research objectives were fulfilled to construct the CROssBAR resource:
- Biomedical data integration: CROssBAR database is constructed by collecting relational data from various biomedical data resources UniProt, IntAct, DrugBank/ChEMBL/PubChem, Reactome/KEGG, OMIM/Orphanet/EFO/HPO by persisting specific data attributes with the implementation of logic rules, in MongoDB collections.
- Deep learning-based predictive models: the main purpose here was to enrich the integrated biomedical data by identifying the unknown interactions between drugs / drug candidate compounds and target proteins. We trained our systems using carefully filtered data in the CROssBAR database, and ran our trained-models on large-scale compound and protein spaces to obtain comprehensive bio-interaction predictions.
- Biomedical knowledge graphs: Different biological components; drugs/compounds, genes/proteins, pathways, phenotypes/diseases are represented as nodes, and their known and predicted relationships are annotated as edges. These intensely-processed heterogeneous biological networks will be utilized to aid biomedical research, especially to infer mechanisms of diseases in relation to biomolecules, systems and candidate drugs.
- Open-access web-service: The aim here is to make the CROssBAR data available to the public in an easily interpretable, interactive way. Knowledge graphs are presented visually on web-browsers as Cytoscape networks. Users can make searches with CROssBAR components, individually or in combination, to obtain relevant sub-networks, which is constructed on-the-fly.
- Experimental validation: In vitro cell based wet-lab experiments are conducted based on the computationally-inferred information, with the purposes of verifying the predictions and also for gaining biological insight in the framework of health and disease, especially to make a contribution to the understanding of processes active in certain cancer subtypes.
For more details about the project please read CROssBAR article or visit our project GitHub repository at: https://github.com/cansyl/CROssBAR and our project website at: http://cansyl.metu.edu.tr/crossbar
Schematic Representation of the CROssBAR project
CROssBAR Team:
Comprehensive Resource of Biomedical Relations with Deep Learning and Network Representations (CROssBAR) is a joint research project between the Middle East Technical University (METU) and Hacettepe University, Turkey, and the European Bioinformatics Institute (EMBL-EBI), UK. The project is funded by the Scientific and Technological Research Council of Turkey (TUBITAK) and the British Council. The people working on the project is given below:
